A group of engineers from Stanford University developed a new laser technique that can detect and identify bacteria in fluids such as blood and wastewater in just minutes.
The research team used an old inkjet printer that utilizes acoustic pulses and achieved a faster, cheaper, and more accurate way to spot bacteria in any fluid sample one might want to test for microbes.
"We can find out not just that bacteria are present, but specifically which bacteria are in the sample – E. coli, Staphylococcus, Streptococcus, Salmonella, anthrax, and more," said Jennifer Dionne, an associate professor of materials science and engineering and, by courtesy, of radiology at Stanford University, in an institutional press release.
"Every microbe has its own unique optical fingerprint. It’s like the genetic and proteomic code scribbled in light."
Currently-used culturing methods take several hours or even days. A tuberculosis culture, for example, takes 40 days, according to Dionne. On the other hand, the new technique can be done in minutes, promising a more accurate and faster way of diagnosing infections. This could pave the way for improved antibiotic usage, safer foods, improved environmental monitoring, and faster medication development.
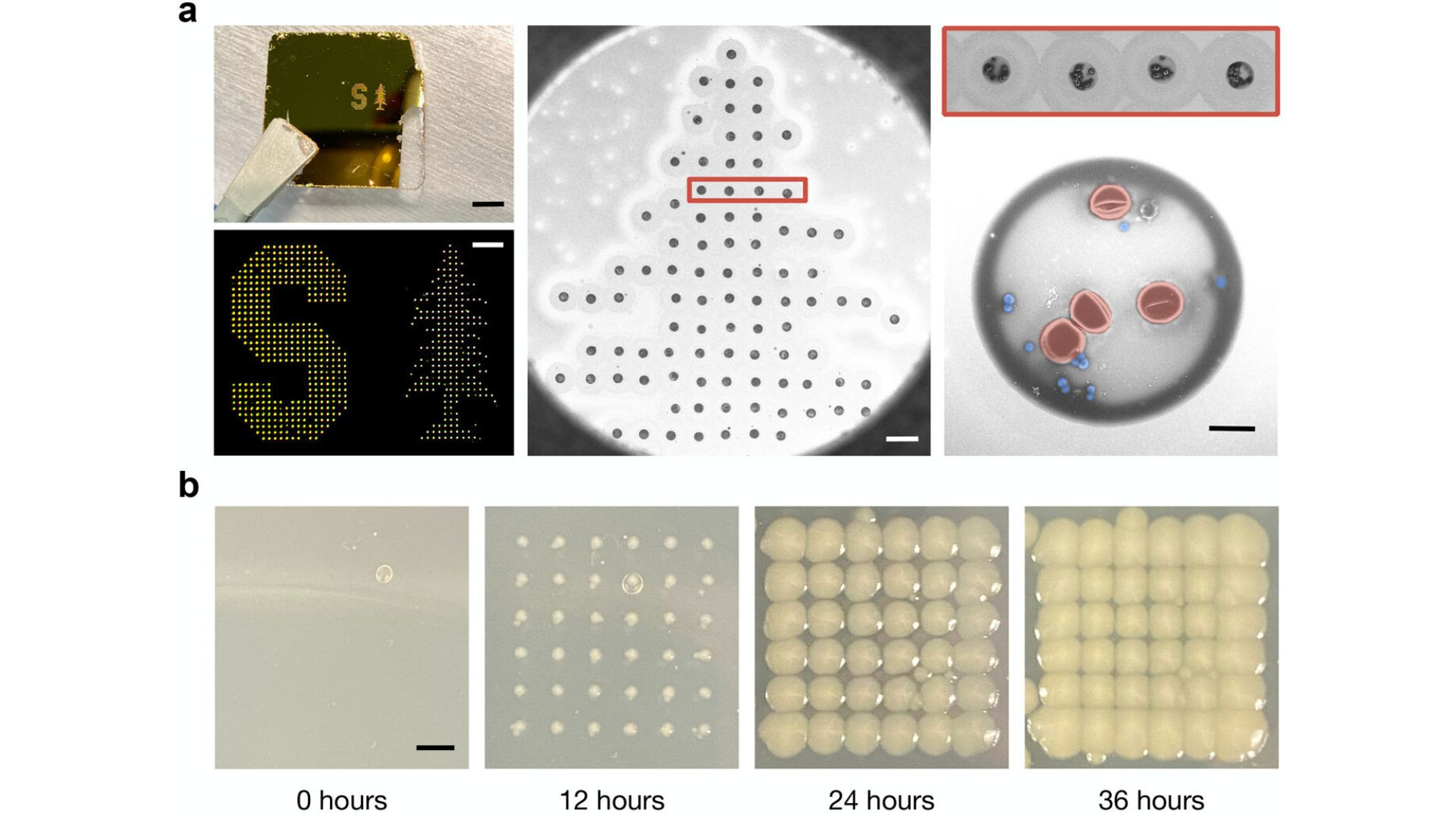
Incorporating a modified inkjet printer and machine-learning algorithms
The team incorporated a modified inkjet printer that prints tiny liquid dots on a slide that are two trillionths of a liter in volume and contain only a few dozen cells, making it easier to see any bacteria present. Gold nanorods added to the sample enhance the bacteria's reflected spectral fingerprint by 1,500 times, making it easy for software to identify the type of bacteria.
“Not only does each type of bacterium demonstrate unique patterns of light, but virtually every other molecule or cell in a given sample does, too,” said first author Fareeha Safir, a Ph.D. student in Dionne’s lab.
“Red blood cells, white blood cells, and other components in the sample are sending back their own signals, making it hard if not impossible to distinguish the microbial patterns from the noise of other cells.”
The final step was to use machine learning to compare the various spectra reflecting from each printed dot of fluid to identify any bacteria in the sample.
Fareeha Safir
“It’s an innovative solution with the potential for life-saving impact. We are now excited for commercialization opportunities that can help redefine the standard of bacterial detection and single-cell characterization,” said senior co-author Amr Saleh, a former postdoctoral scholar in Dionne’s lab and now a professor at Cairo University.
The study was published in the journal Nano Letters.




